Shi lab advancing spatial transcriptomics with custom microscope
OCTOBER 2025
Hailing Shi and her lab are advancing a powerful technique called spatial transcriptomics. It combines conventional information about gene expression (what genes are active) with spatial information: the cell type, how the cells are arranged and even what part of the cell.
Conventional gene expression studies – whether in bulk or single-cell – require tissue dissociation or homogenization. Spatial transcriptomics retains 3D structure, allowing researchers to see the arrangements of cell types and how gradients influence gene expression. With spatial transcriptomics, cancer researchers can distinguish cancer cells from immune cells or stromal cells within the tumor microenvironment, and Identify niches of drug resistance or immune evasion. Neuroscientists can map the signatures of various types of neurons and glia in specific brain layers or circuits. Developmental biologists can track how cell identity and gene expression change across tissue layers during embryogenesis.
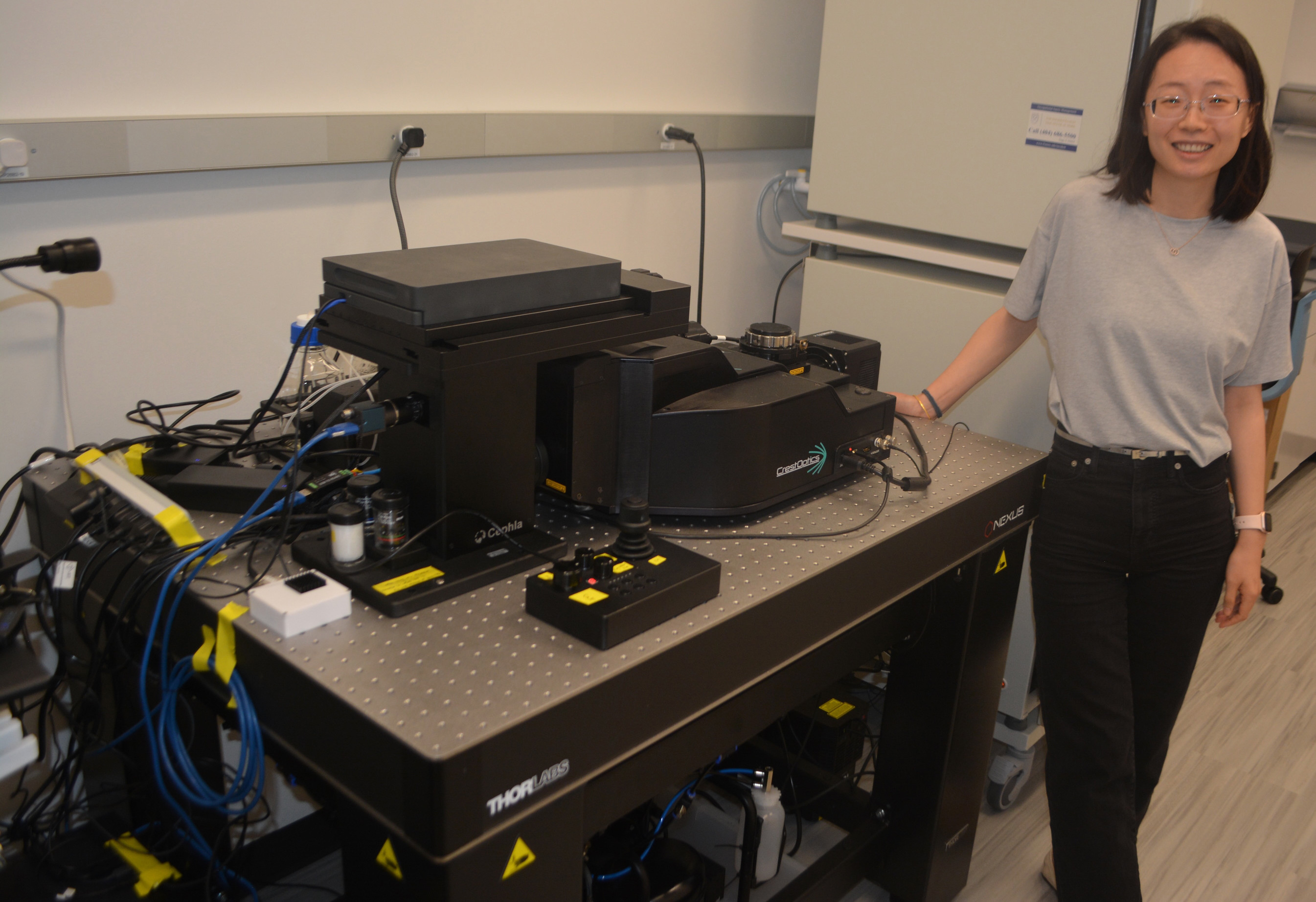
To drive their research, Shi’s lab recently installed a customized microscope set-up in HSRB2. The set-up includes a spinning disc confocal microscope, a high-power camera, a five-color laser, and programmable imaging acquisition software. The water immersion objective lens of the system comes with a programmable water dispenser, allowing for extended imaging sessions lasting for days.

